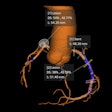
Dynamic PET measures the time-dependent spatial distribution of tracers in blood and tissues, enabling quantification of physiological parameters at the individual voxel level. But though many clinical PET protocols now involve whole-body acquisitions using multiple bed positions, the complexity of dynamic PET data acquisition means that such scans have been constrained to single bed fields-of-view. Whole-body dynamic PET has not yet been employed in routine clinical imaging.
To address this challenge, a U.S.-Swiss research team has developed a parametric imaging method for whole-body dynamic PET. Their dynamic PET parametric imaging framework involves four to six whole-body passes, with whole-body PET images first reconstructed using a 3D maximum-likelihood expectation maximization (ML-EM) algorithm, followed by linear or nonlinear Patlak analysis (a class of graphical techniques for analyzing tracer pharmacokinetics) employed on a voxel level to estimate parametric Patlak images. The proposed framework enables whole-body parametric Patlak imaging for the cost-efficient commercial PET systems of limited axial field-of-view that are now established in the clinic.
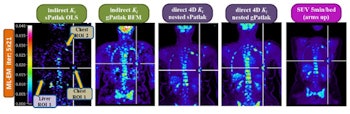
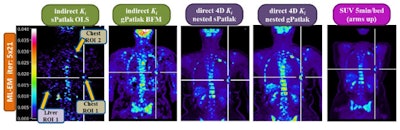
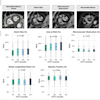
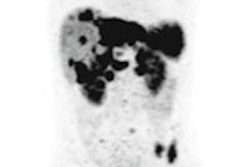
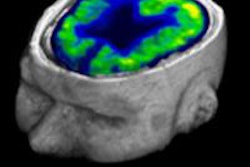
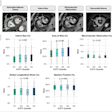
Clinical whole-body (s/g) Patlak Ki images, as estimated either indirectly or directly from the raw dynamic PET data with 4D and indirect methods with patient arms at the bottom position to withstand longer scan duration. Also shown is the respective static SUV image after repositioning same patient with arms in the standard upper position.
This two-step technique represents an "indirect" parametric imaging method. In their latest work, the researchers propose methods for reconstructing whole-body parametric PET images directly from the raw multi-pass PET data to efficiently suppress the statistical noise levels, by employing a 4D reconstruction algorithm (Physics in Medicine and Biology, August 2016, Vol. 61:15, pp. 5456-5485).
"The systematic and integrated use of dynamic PET data offered by direct Patlak methods leads to enhanced quantification in the final whole-body images with clinically acceptable noise levels," explained author Nicolas A. Karakatsanis, from Icahn School of Medicine at Mount Sinai in New York. "Thus, direct Patlak whole-body imaging facilitates our most important aim -- clinical translation of advanced quantitative PET for added value beyond currently established standardized uptake value [SUV] PET images."
To assess the performance of the Patlak algorithms, Karakatsanis, together with Habib Zaidi (University of Geneva), Arman Rahmim (Johns Hopkins University), and colleagues, simulated emission images by applying realistic time-activity curves (derived from published F-18 FDG tracer kinetic parameters) to a "voxelized" XCAT human torso digital phantom. They added six spherical tumor lesions, three in the liver and three in the right lung. Noise-free and noisy dynamic PET projection data were generated and reconstructed to create dynamic PET and Patlak parametric images of the tracer influx rate constant Ki.
The researchers examined both standard Patlak (sPatlak) imaging and the generalized Patlak (gPatlak) method, which also accounts for tracer uptake reversibility. For noise-free reconstructions, indirect and direct (s/g) Patlak Ki images exhibited a higher target-to-background ratio (TBR) than SUV images, for all evaluated lesions.
In noisy Ki images, the researchers observed significantly reduced noise at matched resolution (and vice-versa) for direct versus indirect Patlak methods, especially for regions of low uptake. These findings were supported by quantitative analysis of TBR and contrast-to-noise ratio (CNR).
In general, gPatlak algorithms provided more accurate Ki estimates than sPatlak counterparts. For lung tumors, which had a higher degree of uptake reversibility, both noise-free and noisy gPatlak reconstructions yielded higher Ki TBR than respective sPatlak reconstructions, demonstrating the advantage of gPatlak in regions with nonnegligible uptake reversibility. However, the data indicated higher Ki image noise for gPatlak than sPatlak.
 Nicolas Karakatsanis displays the PET/CT scanner.
Nicolas Karakatsanis displays the PET/CT scanner.The team has also designed a novel nested ML-EM gPatlak reconstruction algorithm, which accelerates convergence by nesting the fast image-based estimation of Patlak parameters within each iteration of the slower projection-based estimation of dynamic PET images. Comparing conventional versus nested direct Patlak algorithms, they observed faster convergence for the nested algorithms. As gPatlak optimization is more susceptible to noise than sPatlak, they proposed initializing the gPatlak algorithm with estimates derived from a few sPatlak iterations. This approach facilitated convergence and resulted in higher Ki lesion contrast.
Clinical validity
Finally, the researchers reconstructed five clinical whole-body dynamic datasets. Using patient data recorded 10 to 45 min postinjection, they successfully produced quantitative whole-body parametric PET clinical images of high contrast and low statistical noise in identified tumor lesion regions.
Background noise levels in direct (s/g) Patlak Ki clinical images were comparable to static image noise in respective whole-body SUV images acquired 60 min postinjection. TBR and CNR for liver and chest regions were higher for Ki imaging than SUV.
"After employing a clinically feasible PET scan protocol and a single 4D reconstruction algorithm, the contrast-to-noise ratio performance was significantly enhanced," Karakatsanis said. "This improves lesion detectability and quantification beyond what is currently established in clinical SUV PET imaging."
The researchers observed lower noise for direct relative to indirect Patlak methods, as well as superior TBR and CNR performance for all direct Patlak methods compared with respective indirect methods. Results also demonstrated superior TBR scores for nested over conventional Patlak Ki images. Despite higher noise levels in gPatlak Ki images, these exhibited the highest clinical CNR scores. Improved TBR and CNR were seen in clinical gPatlak Ki images when at least three cycles of 21 ML-EM sPatlak iterations were employed for its initialization.
"This study demonstrated the clinical feasibility and importance of extending direct 4D Patlak algorithms from single-bed to whole-body PET acquisitions," Karakatsanis told medicalphysicsweb. "I am currently leading a collaboration between Icahn School of Medicine at Mount Sinai, Johns Hopkins University, Geneva University Hospital, and Siemens Molecular Imaging to develop a simultaneous whole-body SUV/Patlak-4D imaging framework capable of producing both static SUV PET images and quantitative Patlak-4D images from a single dynamic whole-body PET scan, performed within the period employed for standard-of-care SUV PET scans."
© IOP Publishing Limited. Republished with permission from medicalphysicsweb, a community website covering fundamental research and emerging technologies in medical imaging and radiation therapy.