An algorithm that generates reproducible, personalized maps of neural networks in the brain using functional MRI (fMRI) has been demonstrated by an international collaboration. The noninvasive approach could enable more efficient planning of neurosurgery for brain tumors and disorders such as epilepsy. Other potential applications include the guidance of brain stimulation techniques used to treat neurological and psychiatric conditions.
Functional networks in the cerebral cortex, like those responsible for speech and decision making, vary significantly between individuals. Consequently, neurosurgeons must map them prior to surgery to limit damage. However, the gold standard means of doing this -- cortical stimulation -- requires the direct placement of electrodes on the brain, and to date there has been no viable noninvasive alternative.
 Hesheng Liu, PhD, and Dr. Danhong Wang.
Hesheng Liu, PhD, and Dr. Danhong Wang."fMRI has already been used for presurgical mapping, but with limited success due to its low reliability," said first author Dr. Danhong Wang, a research fellow in radiology at the Athinoula A Martinos Center for Biomedical Imaging at Massachusetts General Hospital and Harvard Medical School. "We believe a more reliable and accurate mapping technology will greatly expand the clinical use of fMRI."
The research was led by Hesheng Liu, PhD, director of the Laboratory for the Study of the Brain Basis of Individual Differences (Nature Neuroscience, Vol. 18:12, pp. 1853-1860).
Functional mapping of the brain is an active area of research and so-called "parcellating" algorithms are under development by several groups. However, the collaboration's iterative algorithm is unique in that it exploits a priori knowledge of functional brain anatomy. It uses an atlas of 18 brain networks as a template and starting point, projecting them onto the individual's blood oxygen level dependent (BOLD) fMRI data. The atlas was derived from the scans of 1,000 individuals in previously reported work.
The parcellation algorithm compares each of 18 network-averaged reference signals derived from the template with fMRI data from single points in the individual's scan. Points that correlate strongly with any of the 18 reference signals are averaged to derive more personalized "core" signals for each network.
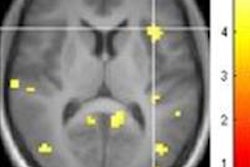
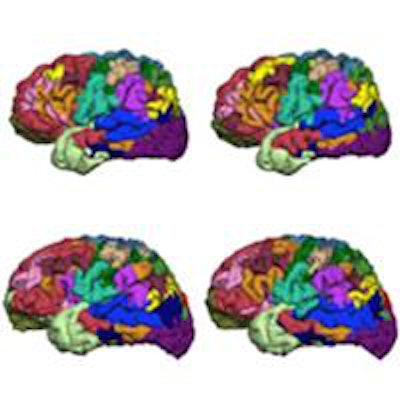
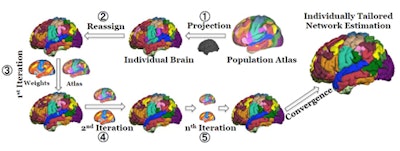 The iterative parcellation algorithm incorporates a functional atlas of the cerebral cortex generated from fMRI scans of 1,000 volunteers and data on intrasubject variability to map 18 functional networks.
The iterative parcellation algorithm incorporates a functional atlas of the cerebral cortex generated from fMRI scans of 1,000 volunteers and data on intrasubject variability to map 18 functional networks.Reference and core signals are then iteratively averaged together to update the reference signal, eventually converging to a final, personalized set of networks. Each time they are averaged, the core signal is preferentially weighted depending on several factors. For example, core signals are assigned a greater weight than the atlas-derived reference signals at anatomical locations known to have greater inter-subject variability, based on data previously reported by Liu's group.
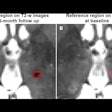
The collaboration confirmed the algorithm's reproducibility and that it could detect differences between individuals in two fMRI datasets of 25 and 100 volunteers. In the first set, for example, where subjects were scanned five times over six months, intrasubject reproducibility quantified using a Dice coefficient was 83%. Analysis of the second set also demonstrated reproducibility when the algorithm was applied to resting state scans and concatenated scans where the volunteers were given tasks to activate specific networks.
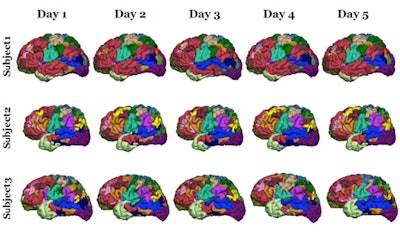 A study of 23 healthy volunteers scanned five times over six months revealed high intrasubject reproducibility and demonstrated that the parcellating algorithm could detect intersubject variation.
A study of 23 healthy volunteers scanned five times over six months revealed high intrasubject reproducibility and demonstrated that the parcellating algorithm could detect intersubject variation.The researchers validated the algorithm against invasive cortical stimulation in eight presurgical epilepsy patients and compared its performance to other parcellation methods. They included simple application of the population-defined networks used by the algorithm and conventional task-activated fMRI scans where networks are identified using statistical techniques.
Algorithm results were consistent with the cortical stimulation data, providing a closer match than the other methods, quantified using receiver operating characteristic curves. For example, the algorithm gave an area under the curve of 0.91 compared with 0.76 for task activation fMRI.
"We have shown that sensory and motor networks can be accurately mapped in a few patients prior to brain surgery," Liu said. "We must [now] validate the localization of cognitive networks, such as the language networks, using invasive measures in a large group of patients."
The collaboration hopes to eventually replace invasive cortical stimulation with this technique. "My optimistic expectation is that a noninvasive functional network parcellation will be used routinely in the clinic, at least as a prescreening technique for invasive tests, within five years," Liu told medicalphysicsweb.
Jude Dineley is a freelance science writer and former medical physicist based in Sydney, Australia.
© IOP Publishing Limited. Republished with permission from medicalphysicsweb, a community website covering fundamental research and emerging technologies in medical imaging and radiation therapy.