The development of automated image segmentation schemes can be enhanced significantly with the application of a process called transfer learning, according to a new Dutch study published in the May edition of IEEE Transactions on Medical Imaging.
The technique's advantage lies in its ability to cope with differences in data distribution between training and target data. It can lead to improved performance over supervised learning for segmentation across scanners and scan protocols.
 Annegreet van Opbroek from Erasmus University Medical Center.
Annegreet van Opbroek from Erasmus University Medical Center."We showed that transfer learning can be helpful for segmentation of images from different scanners, imaging protocols, and patient groups," wrote lead author Annegreet van Opbroek, a doctoral student at Erasmus University Medical Center in Rotterdam, in an e-mail to AuntMinnieEurope.com. "Transfer learning has been around in the machine-learning community for quite some time, and various interesting algorithms have been developed, but it is only very recently getting some attention in medical image segmentation."
The study showed that even with little representative training data available, transfer learning greatly outperformed typical supervised learning algorithm approaches, reducing classification errors "by as much as 60%," van Opbroek and colleagues wrote (IEEE Trans Med Imaging, May 2015, Vol. 34:5, pp. 1018-1030).
A new approach
Data segmentation is required for medical procedures and diagnoses that encompass everything from transplantation to surgery. But the process is hamstrung by the need for customization and extensive training of datasets for each possible variation.
"The variation between images obtained with different scanners or different imaging protocols presents a major challenge in automatic segmentation of biomedical images," they wrote.
The investigators built their new classification model to deal with the longstanding problem of customizing segmentation algorithms.
Transfer learning is a new form of machine learning that allows for differences between training and target domains, the authors explained. The algorithms "exploit similarities between different classification problems or datasets to facilitate the construction of a new classification model," they wrote. "They possess the ability of supervised-learning algorithms to capture class-specific knowledge in the training phase without requiring exactly representative training data."
Extra data from different sources
Classification data can be dissimilar but still useful for creating a classification model. It enables the training and test data to follow different distributions, possess different labeling functions, and even consist of different data classes, the study authors wrote.
Data that follow the same distribution and have the same labeling function and features are referred to as data from the same source. It is the model for conventional supervised-learning algorithms.
Transfer learning's goal, on the other hand, is to learn a classification algorithm for the target data that benefits from already available data originating from different sources. These data are somewhat similar, but not completely representative, of the target data.
Previous articles have distinguished between three types of transfer learning: inductive transfer learning, where the transfer and target data can have different labeling functions, different features, and sometimes prior distributions.
"We assume that a small number of labeled training samples from the target source is available, the so-called same-distribution training data, and aim to transfer knowledge from a much larger amount of labeled training data that is available from sources other than the target data, the so-called different-distribution training data," the authors wrote.
Despite varying label functions between training and target sources, inductive transfer learning still assumes that different-source data are similar enough to provide some extra information within the feature space when same-source data are scarce, they added.
4 classifiers
"The main goal of this paper was to draw attention to the fact that transfer-learning algorithms can be used to train on data that originate from different scanners/imaging protocols/patient groups than the test data," van Opbroek wrote.
The study presents four transfer classifiers that use this same- and different-distribution training data, all of which are based on support vector machine (SVM) classification.
Three of the four classifiers used sample weighting, in which both the same- and different-distribution training samples are used for training, but the different-distribution samples are given lower weight.
- The first is called weighted SVM, for which both same- and different-distribution training samples are used for training, the latter with a lower weight than the former.
- The second is called reweighted SVM, an extension of the SVM in which the weights of misclassified different distribution training samples are reduced iteratively.
- The third is called TrAdaBoost, which builds a boosting classifier for transfer learning by increasing the weight of misclassified same-distribution samples while reducing the weights of misclassified different-distribution samples.
- The fourth, adaptive SVM, is not based on sample weighting. Rather, it trains an SVM on the same-distribution samples only.
The experiments focused on segmentation through voxelwise classification on data from several sources acquired using different MRI systems.
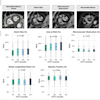
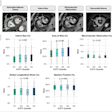
The study looked at segmentation of white and gray matter from cerebrospinal fluid (WM/GM/CSF), and distinguished white matter from multiple sclerosis lesions, in each case comparing the performance of the four transfer classifiers to that of two conventional supervised-learning classifiers, including a regular SVM trained on every training sample, and an SVM trained on same-distribution training samples.
Transfer learning always better
The four transfer classifiers were weighted SVM (WSVM), reweighted SVM (RSVM), TrAdaBoost, and adaptive SVM (A-SVM).
- WSVM proved the most consistent classifier of the four; and with very few transfer learning samples outperformed the regular SVMs on all training data and learning curves.
- RSVM performed similarly to WSVM on the lesion segmentation experiments, but fell short of WSVM's performance on the WM/GM/CSF segmentation experiments.
- TrAdaBoost was the least helpful, never outperforming the two baseline supervised-learning classifiers. TrAdaBoost did especially poorly with lesion segmentation, where classification errors increased as training samples were added.
- The performance of A-SVM was dependent on the classification task. A-SVM performed very well on the WM/GM/CSF segmentation experiments when more than 15 training samples were available but poorly on the lesion-segmentation experiments.
Broad applicability
Even when very few training samples were available, "transfer learning greatly outperformed the supervised-learning classifiers, minimizing mean classification errors by up to 60%," the team wrote.
"Transfer-learning applied to segmentation enables supervised segmentation of images acquired with different MRI scanners or imaging protocols," the group wrote.
And while the study focused on brain segmentation, variability is a common problem across many applications, so transfer learning has broad applicability.
"We believe that transfer learning is a promising approach to biomedical image analysis," van Opbroek and colleagues wrote. In applications where ground truth labels are available from other studies, "transfer learning can significantly decrease the amount of representative training data needed."
There are many more applications where a model is learned from labeled training samples, such as segmentation of organs (e.g., liver, prostate), brain structures, and tissues (e.g., carotid plaque, knee cartilage, tumors), but transfer learning can also be interesting in other areas of medical image analysis, such as computer-aided diagnosis, they concluded.