Texture analysis may still be experimental, but it's becoming a leading cancer research technique in several institutions around the world. Technology developed in the U.K. is being applied to MR and CT images to predict the aggressiveness and prognosis of cancers throughout the body including the lungs, esophagus, head and neck, prostate, and most recently, colon and rectum.
In one of several studies to be presented at the upcoming 2014 RSNA meeting next week in Chicago, the algorithm developed at the University of Sussex in the U.K. will demonstrate that the tool, known as TexRAD, can help hospitals make earlier and more accurate treatment decisions and survival assessments for patients with colorectal cancer.
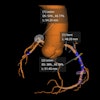
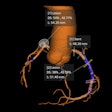
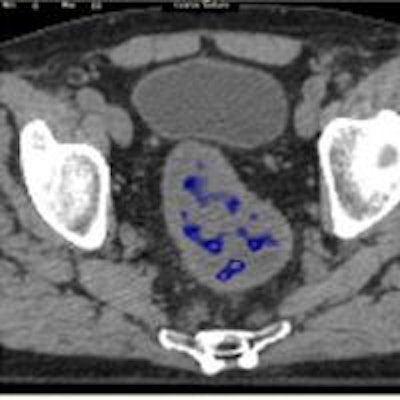
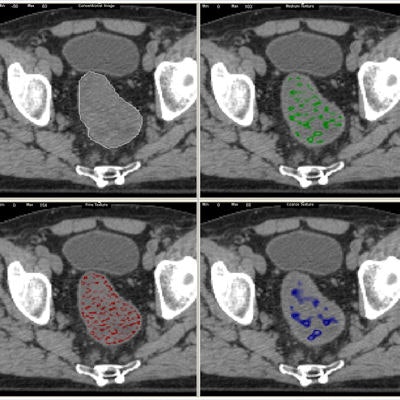 A standard CT scan (top left) with the tumor outlined in white. With the TexRAD software applied, it is possible to analyze the tumor's texture at a fine (red), medium (green), and coarse (blue) level. Image courtesy of bowel cancer case analysis from University College London Hospital.
A standard CT scan (top left) with the tumor outlined in white. With the TexRAD software applied, it is possible to analyze the tumor's texture at a fine (red), medium (green), and coarse (blue) level. Image courtesy of bowel cancer case analysis from University College London Hospital.Colorectal cancer kills more than 16,000 people a year in the U.K., making it the second-most common cause of cancer death after lung cancer.
In one study, texture analysis was found to enable early diagnosis of those bowel cancer patients who weren't responding to standard cancer therapies better than other available tumor markers. The same TexRAD markers also showed the ability to assess at an early stage the likelihood of survival, distinguishing patients who will have a good prognosis from those having poor prognosis, the authors report.
Balaji Ganeshan, PhD, who initiated research into the technique as part of his PhD in biomedical engineering in 2004, said by using TexRAD to scan for subtle anomalies in a tumor's texture, researchers have been able to spot more quickly when treatments are -- or are not -- working and adjust treatment accordingly. And because it provides an extra layer software analysis to MRI and CT scans that already exist as part of routine clinical practice, it is noninvasive from the patient's point of view and potentially cost-effective to the healthcare provider.
"You might have two kinds of patients with the same tumor type, but they may have very different outcomes -- one might do really well and the other, not," he told AuntMinnieEurope.com by telephone. "You identify early who is going to do well and who is not, then you change the treatment accordingly for better patient outcomes and that's the key word and that's where texture analysis comes in."
There are several advantages to this noninvasive approach, he said. For one thing, advanced imaging is expensive, so it's critical to extract the most information possible from existing scans. "You don't have to do any extra scans -- that's the beauty of it," he said.
And because cancer patients normally have scans at different time points during treatment and follow-up, there is plenty of data to study, he said, "a plus for the patient and a plus for the facility, because ultimately it's a software analysis."
As soon as it became clear the tool worked, it was time to seal it up, Ganeshan recalled. "I completed the PhD research in 2007 and by that time we had applied for patents," he said. "We got several in Europe, plus Japan, and Canada -- U.S. is a pending patent."
The technology is also approved for research in the U.S. and Europe, and commercial marketing rights including a 510(k) in the U.S. is expected in 2015. Meanwhile, TexRad is being evaluated in several leading research institutions and university hospitals around the world including Johns Hopkins University in Baltimore, the University of Mississippi in Jackson, and European centers including its academic home at University College London, as well as the universities of Brighton and Sussex in the U.K., the University of Rome in Italy, and the University of Basel in Switzerland.
"We have quite a number of sites and a lot of momentum and interest," Ganeshan said. "It's fair to say we've been one of the pioneers."
Upcoming presentations
In two U.K. studies to be presented at RSNA 2014, researchers analyzed the tumors of colorectal cancer patients.
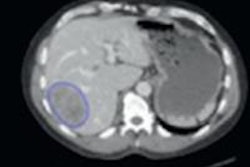
In a series from University College London Hospitals (UCL), for example, researchers analyzed baseline PET/CT scans and then followed up patients for an average of three years. The tumor image analysis enabled them to accurately predict patient survival, Ganeshan, Dr. Ming Young Simon Wan, and colleagues wrote in their RSNA abstract (SSQ06-05).
The image filtration-histogram technique was used to assess the CT images, and the investigators also measured FDG uptake (SUVmax) on PET, determining clinical stage using surgical histology and imaging data.
The research team grouped the patients by stage: stage I-III rectal cancer, stage I-III colon cancer, and metastatic stage IV cancer colorectal cancer. Median follow-up for surviving patients was 47.9 months (minimum 12 months). For patients with stage I-III rectal cancer (n = 42), CT texture analysis (coarse skewness, p = 0.011), SUVmax (p = 0.012) and clinical stage (p = 0.006) were the best predictors of survival.
The only independent predictor of survival (p = 0.003) was a significant interaction between skewness and clinical stage. For patients with stage I-II and stage III cancers, texture analysis (unfiltered kurtosis, p = 0.001) and T stage were the only significant survival predictors. And for stage IVb disease, texture analysis (fine kurtosis) was again the only significant predictor of survival.
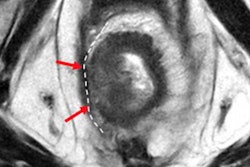
Another study to be presented at RSNA (SSA07-04) looks at tumor heterogeneity in MRI as a potential prognostic biomarker in patients with rectal cancer who have undergone chemoradiotherapy.
"It's one thing to determine if it's a tumor or not, but we have taken it a step further to determine the prognosis where imaging wasn't traditionally used as much," Ganeshan said.
A third RSNA presentation by Ganeshan and colleagues (SSA07-06) looks at the performance of texture analysis, diffusion-weighted imaging (DWI), and perfusion imaging in predicting treatment response in rectal cancer patients studied with 3-tesla MRI.
Behind the software
The underlying principle is the same for CT and MRI analysis: It's simply a matter of tuning the software to Hounsfield Units or signal intensity. The software technique approach starts with a filtration histogram analysis; the second step is quantification. The filtration extracts features of different sizes and variations in intensity from the tumor, enhancing and quantifying these features, he explained.
"What happens in routine practice is that the radiologist looks at the tumor and reports whether it is heterogeneous or homogeneous, but it's very subjective," Ganeshan noted. "One may think it is heterogeneous and another might think it is more homogenous, so it depends on the radiologist's experience, and the human visual system has some limitations in how you can actually see patterns."
TexRad breaks down tumor images into different components separately -- fine structures separately, medium structures separately, and the large coarser structures separately, and it breaks down the images and quantifies the different distributions or patterns via the histogram analysis.
"And the different parameters that come out of it can tell you about the different aspects of heterogeneity," he said. "The combination of histogram approach filtration and the histogram together make it a powerful approach.
In analyzing the textures to try to predict the aggressiveness of a lesion, "It's actually emulating what radiologists are trying to do but giving objectivity to the practice rather than subjectivity," he said. Only using the software, time after time, the same ROI, and the same region of interest will get the same numbers and the same marker of heterogeneity.
Lately, "We're doing more research in terms of correlating the results with genetic mutations and genomes and so on, so it's a fascinating era for this area," he said.