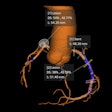
Accurate fully automatic segmentation of subcutaneous fat, visceral fat, and psoas muscle from abdominal CT is feasible without human input. That's the main finding of new artificial intelligence (AI) research conducted at the Utrecht University Medical Center (UMC) in the Netherlands.
"The amount and anatomical distribution of fat and muscle in different body compartments is an important prognostic factor in patients with cardiovascular disease," explained R&D engineer Pim Moeskops, PhD, and colleagues at the UMC Utrecht. "Although this information is routinely contained in many types of CT scans, it is hard to quantify in daily clinical routine because manual segmentation is time-consuming, especially in 3D."
The group has investigated the use of a deep learning-based method for automatic segmentation of subcutaneous fat, visceral fat, and psoas muscle, unveiling the findings at the European Society of Cardiovascular Radiology (ESCR) annual congress, held in Antwerp, Belgium, from 24 to 26 October.
The researchers evaluated the viability of fully automated body composition measurement using a dataset of 20 CT scans of the abdomen (in-plane resolution, 0.63 mm to 0.75 mm; slice thickness, 5 mm; slice increment, 5 mm). Trained observers defined the reference standard by manual annotation of subcutaneous fat, visceral fat, and psoas muscle in all slices that contained the psoas muscle.
"Images from 10 patients were used to train a dilated convolutional neural network with a receptive field of 131 × 131 voxels to distinguish between the three tissue classes," the authors explained. "Voxels were assigned to the class with the highest probability. Data from the remaining 10 patients were used to evaluate the performance of the method."
They evaluated segmentation performance with Dice coefficients between the manual and automatic segmentations. Additionally, linear correlation coefficients known as Pearson's r were computed between the manual and automatic segmentation volumes.
On average, segmentation of a full scan took about 15 seconds. The average Dice coefficients over 10 test scans were 0.89 ± 0.022 for subcutaneous fat, 0.92 ± 0.042 for visceral fat, and 0.76 ± 0.052 for psoas muscle.
At the L3 vertebrae level, the average Dice coefficients were 0.92 ± 0.019 for subcutaneous fat, 0.93 ± 0.048 for visceral fat, and 0.87 ± 0.035 for psoas muscle. Pearson's r between the manual and automatic volumes were 0.996 for subcutaneous fat, 0.997 for visceral fat, and 0.941 for psoas muscle, stated Moeskops and colleagues.
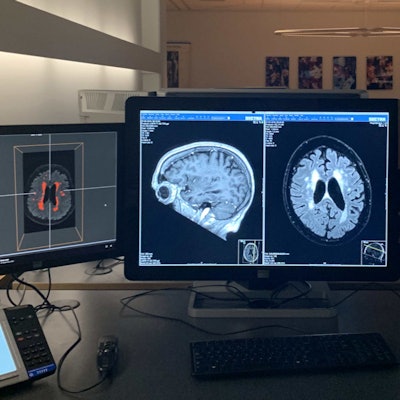
 The IMAGR interface (left monitor) shows the results of the white-matter hyperintensity detection and quantification algorithm. The middle monitor shows the PACS interface. The right monitor shows the electronic health record. Image courtesy of Olivier Middendorp - NRC.
The IMAGR interface (left monitor) shows the results of the white-matter hyperintensity detection and quantification algorithm. The middle monitor shows the PACS interface. The right monitor shows the electronic health record. Image courtesy of Olivier Middendorp - NRC.Instead of using one-off AI solutions for different systems and modalities, the UMC Utrecht team has built a vendor-neutral infrastructure that supports the entire imaging workflow chain, covering patient selection, image planning, acquisition, reconstruction, analysis, reporting, and prognosis. The AI system can "listen in" to hospital information streams and then automatically select an appropriate algorithm to be used.
The AI infrastructure took three years to build and went operational earlier this year. The first application was effective in January 2019, and the technology has been in routine use since August. The project leaders received an initial 150,000 euros from UMC Utrecht, and university managers recently gave them another grant of 620,000 euros.
Moeskops started work as an R&D engineer at medical image quantification software developer Quantib in September 2018. He was the first employee of Quantib-U, the company's joint venture with UMC Utrecht. Quantib's other clinical partners are Erasmus Medical Center and Amsterdam University Medical Center.
He studied biomedical engineering at Eindhoven University of Technology and focused on medical image analysis during his master's studies. While obtaining his doctorate at UMC Utrecht, he specialized in automatic MRI-based quantification of brain characteristics in preterm newborns. His work at Quantib enables him to use his expertise in deep learning for medical imaging to develop practical radiology software that will be used in hospitals, he said in his bio on the company website.
If you'd like to see the authors' full range of clinical images and figures shown at ESCR 2019, you can view their presentation on the Electronic Presentation Online System (EPOS) section of the European Society of Radiology website.