Researchers from the Riken Center for Advanced Intelligence Project (AIP) in Japan have shown that a deep-learning algorithm can be used to extract interpretable features from annotation-free histopathology images from prostate cancer patients. Their framework outperformed the prediction of biochemical recurrence using conventional, Gleason score-based methods.
Prostate cancer is the second most common cancer affecting men worldwide, with an incidence rate of 13.5%, according to the World Health Organization. Expert pathologists diagnose this type of cancer through a transrectal biopsy, following the results of a prostate-specific antigen (PSA) test. The extracted samples of tissue are examined under a microscope and, if cancerous cells are found, divided into risk groups assigned through the Gleason score. This grading system is considered the gold standard in cancer medicine, as it determines the aggressiveness of prostate cancer and helps doctors establish the right course of treatment.
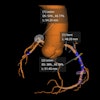
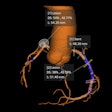
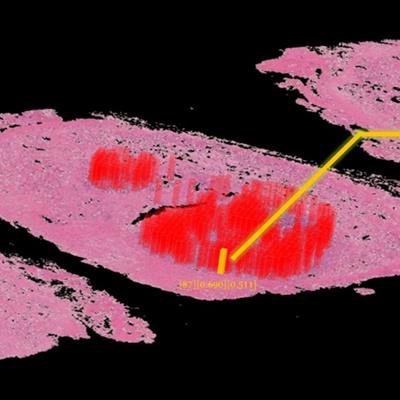
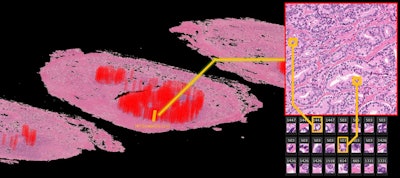 The deep-learning algorithm automatically annotates a 3D whole-mount pathology image and presents the discovered features in an understandable way. The patches with high probability of cancer recurrence are shown by color and height. Image courtesy of Riken.
The deep-learning algorithm automatically annotates a 3D whole-mount pathology image and presents the discovered features in an understandable way. The patches with high probability of cancer recurrence are shown by color and height. Image courtesy of Riken.This type of diagnostic pathology, however, requires expert knowledge, is time-consuming, and can suffer from interobserver variability. Even though automated machine-learning tools capable of accurately classifying histopathology images exist, these methods have not yet gained clinical approval -- mainly because deep-learning algorithms suffer from a lack of interpretability, making their decisions hard to visualize or even explain.
AI-generated features
Paving the way toward interpretable clinical analyses, the investigators developed an artificial intelligence (AI) framework capable of acquiring interpretable features from annotation-free histopathological images. To achieve this, they used whole-mount pathology images acquired at three different centers. This included images from 842 patients at Nippon Medical School Hospital (NMSH), plus 95 patients from St. Marianna University Hospital (SMH) and Aichi Medical University Hospital (AMH) (Nature Communications, 18 December 2019).
 First author Dr. Yoichiro Yamamoto, PhD.
First author Dr. Yoichiro Yamamoto, PhD.The team used images from 100 patients at NMSH to extract the features, while the rest of the NMSH dataset was used to validate their method. Lead researcher Dr. Yoichiro Yamamoto, PhD, and his colleagues trained two unsupervised deep neural networks, known as deep autoencoders, to reduce the 10-billion-scale pixel data into 100 features. They achieved this by using both low-resolution and high-resolution histopathology image patches, inspired by the diagnostic process of pathologists. The computation was performed on AIP's Riken Alp Deep Learning Environment (RAIDEN) supercomputer.
To validate their work, the researchers used the 100 generated features to predict cancer recurrence in the remaining NMSH dataset. At the same time, they used the human-established cancer criteria, the Gleason score, to make the same predictions. Their results showed that predictions of biochemical recurrence were more accurate with AI-generated features than when using the conventional method.
Moreover, when combining the Gleason score with the machine-generated features, the accuracy further increased. Furthermore, this framework delivered similar accuracies when used with the SMH and AMH datasets, revealing the potential for broad use.
Finally, the authors evaluated the AI-generated features. They retrieved the most representative images for each of the features and asked an expert pathologist to examine them.
"In summary, the pathologist found that the deep neural networks appeared to have mastered the basic concept of the Gleason score fully automatically, generating explainable key features that could be understood by pathologists," they noted.
 The RAIDEN supercomputer. Image courtesy of Riken.
The RAIDEN supercomputer. Image courtesy of Riken.The researchers pointed out that the deep neural networks identified features of stroma in the noncancerous area as prognostic factors, and that such features typically have not been evaluated in prostate histopathological images. This is a key result as it raises the possibility of a tool capable of discovering new and uncharted disease characteristics. As future work, the authors plan to further validate the framework by conducting clinical trials and by applying it to other diseases including rare cancers.
Irina Grigorescu is a doctoral student contributor to Physics World. She works at King's College London, where she uses MRI and deep learning to characterize developmental outcomes in babies born preterm.
© IOP Publishing Limited. Republished with permission from Physics World, a website that helps scientists working in academic and industrial research stay up to date with the latest breakthroughs in physics and interdisciplinary science.