Features extracted from routinely acquired CT images can be used to classify risk among prostate cancer patients. Using a machine-learning method, researchers in the U.K. and the Netherlands have trained prediction models to identify textural and intensity-based features imperceptible to human observers, and to associate them with commonly used measures of disease progression. The technique could complement conventionally used methods and in the long run, it lead to a noninvasive alternative to tissue biopsies for guiding treatment decisions.
Prostate cancer is one of the most common forms of cancer, but the after-diagnosis development of the disease is extremely variable. While some tumors metastasize rapidly, others can remain inert for years. To predict the risk represented by a given tumor, oncologists typically assign a Gleason score based on how a sample of the tumor appears compared with normal prostate tissue.
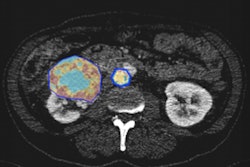
 Sarah Osman, PhD, and colleagues at Queen's University Belfast (QUB) are applying radiomics to CT data from prostate cancer patients. Image courtesy of Kelly Redmond, QUB.
Sarah Osman, PhD, and colleagues at Queen's University Belfast (QUB) are applying radiomics to CT data from prostate cancer patients. Image courtesy of Kelly Redmond, QUB.According to Sarah Osman, PhD, postdoctoral research fellow at Queen's University Belfast (QUB) and Northern Ireland Cancer Centre (NICC), the problem is that "prostate cancer is highly heterogeneous in nature and a limited number of biopsies may not give the full picture." As a complementary method to current biopsy methodologies, Osman -- with collaborators at QUB, NICC, and D-lab at Maastricht University Medical Centre -- has turned to the growing discipline of radiomics.
"Radiomics is the high-throughput extraction of quantitative imaging features with the intent of creating mineable databases from routine medical scans," she explained. "The central hypothesis is that mining of such imaging features will reveal predictive or prognostic associations between images and medical outcomes -- i.e., radiomics features can act as surrogates of biological characteristics."
Although the principle is rooted in image-classification techniques that are decades old, advances in computer hardware and software have prompted an explosion in the field over the last few years. Approaches based on MR images have already shown promise, but as imaging protocols vary between clinics, and MRI scans only recently became part of routine practice for cancer management, it has been difficult to obtain the large datasets needed to test the technique thoroughly.
Instead, Osman and colleagues used CT scans acquired for 342 prostate-cancer patients prior to radiotherapy. Because such images are already a routine part of radiotherapy treatment planning, they are widely available and highly standardized. At the time of treatment, each patient had been assigned a Gleason score according to the results of between six and 21 biopsies, and had been classified as low, medium, or high risk depending upon the size of the tumor and whether the cancer had spread (International Journal of Radiation Oncology, Biology, and Physics, October 2019, Vol. 105:2, pp. 448-456).
Focusing only on the prostate itself, the researchers extracted 1,618 candidate radiomic features from each image. These were based on the statistics and distribution of voxel intensity, and measured different aspects of texture heterogeneity. Of this total, 522 features passed reliability tests and were taken forward for analysis. These features, along with the Gleason score and risk classification for each patient, were used as training data for a machine-learning algorithm.
After training, the classification models proved able to discriminate between patients in low- and high-risk groups, and between those with low and high Gleason score. The system was especially competent at distinguishing between separate patients with the same high Gleason score, but whose biopsies showed subtle morphological differences. This distinction, which is based on the prevalence within the prostate of the most abnormal-looking tissue, has been shown previously to indicate likely disease outcome.
Although the present study will not revolutionize prostate cancer treatment by itself, Osman said, as the first CT-based radiomics investigation for this treatment site, it shows what could be possible in the future. "The idea is that the information gained by radiomics analysis will complement what we get from biopsies, helping us determine risk groups more accurately, and subsequently leading to more personalized treatment decisions."
Marric Stephens is a freelance science writer based in Bristol, U.K.
© IOP Publishing Limited. Republished with permission from Physics World, a website that helps scientists working in academic and industrial research stay up to date with the latest breakthroughs in physics and interdisciplinary science.