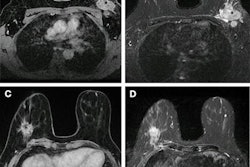
The combination of radiomics and a machine-learning algorithm can be highly accurate for distinguishing between glioblastoma (GBM) and brain metastasis, aiding in the swift diagnosis of brain lesions on standard T1-weighted MRI, according to research published online on 24 June in Physica Medica.
A team of researchers led by Rafael Ortiz-Ramón, PhD, of the Polytechnic University of Valencia in Spain found that the best-performing machine-learning model yielded an area under the curve of 0.896 from its analysis of 2D texture features on MRI.
"These promising results indicate that in the near future radiologists could provide a correct diagnostic with the combination of clinical, radiological and textural information derived from the first MRI evaluation where the lesion is detected," the authors wrote. "This way, patients could avoid invasive and exhaustive additional procedures and will be correctly diagnosed in the earliest stages of the disease."
Accelerating diagnosis
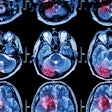
GBM and brain metastases have similar clinical symptoms and appearance on conventional MRI, so a histopathological analysis is currently the only accepted method for a definitive diagnosis. In hopes of developing a decision-support tool to help differentiate these lesions on conventional MRI and speed up diagnosis, the researchers explored the use of radiomics and machine-learning algorithms.
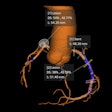
In a retrospective study, the researchers quantified 86 different texture features for segmented lesions in 50 patients with brain metastasis and 50 patients who had received MRI between December 2010 and January 2017. All patients received T1-weighted contrast-enhanced MRI -- including 2D turbo spin echo coronal images -- on an Achieva 1.5-tesla scanner (Philips Healthcare) with an eight-channel SENSE head coil after intravenous administration of a single dose of gadobutrol (Gadovist, Bayer HealthCare).
After lesions were selected and segmented by an expert neuroradiologist with 20 years of experience, the regions of interest were preprocessed to improve texture discrimination.
The researchers then evaluated three feature selection models and five different prediction models.
Using a five-fold cross-validation process, they randomly split the study group into groups of 80 for training and 20 for testing. This process was repeated 10 times.
The best overall performance
Ortiz-Ramón and colleagues found that the highest classification accuracy was achieved using the original images quantified with 128 gray levels and a feature selection method based on the p-value.
| Performance of machine-learning models for classifying GBM and brain metastasis | |||||
| Naive Bayes model | K-nearest neighbors model | Random forest model | Support vector machine (SVM) model | Multilayer perceptron (MLP) model | |
| Area under the curve (AUC) | 0.799 | 0.824 | 0.851 | 0.901 | 0.912 |
Although the MLP model had a slightly higher AUC than the SVM algorithm, the difference was not statistically significant. Furthermore, the SVM algorithm uses only 32 features, almost half as many as the 57 features utilized by the MLP model (57 features). Therefore, the researchers considered the SVM model to have the best overall performance.
"The results presented in this study show that GBMs and [bone metastases] can be classified with a high level of accuracy by employing a set of 2D texture features extracted from structural MRI combined with a machine-learning scheme," the authors wrote.
External validation needed
An external validation of the models would be necessary to check if the generalizability of the models would be limited by the number of the features that are used in the analysis, according to the researchers. In addition, other features such as clinical data or shape descriptors should be included in the models to test their effectiveness and to enhance the radiomics analysis, they said.
"Finally, more patients should be included in the analysis to validate the results using an external cohort and to enable the creation of a final effective predictive model to classify GBMs and [brain metastases]," the authors wrote.