For a while now, relaxation time measurements have returned as diagnostic tools or biomarkers. A quarter of a century ago, scientists and medical doctors had given up such measurements. The early 1990s was the time when the "Relaxation Times Blues" arrived.1 Ian Young, the U.K. medical physicist and one of the leading and most influential scientists in MRI, summed up the trials and errors in a short history of MRI as follows: "Sadly, the many attempts that were made to correlate pathology and relaxation behavior have yielded none of the precise numerical relationships that were hoped for in the early days of MRI, so that this line of investigation ... has now been abandoned."2
A grant-creating perpetuum mobile?
It is rare that a method appears, disappears, and then reappears again, as is the case of tissue characterization based on relaxation time constants. Yet these obsolete methods were dug out again, and research grants were given to answer questions that had been discarded earlier.3,4 New pulse sequences and algorithms were developed, and researchers tried their luck again.
 Dr. Peter Rinck, PhD, is a professor of diagnostic imaging and the president of the Council of the Round Table Foundation (TRTF) and European Magnetic Resonance Forum (EMRF).
Dr. Peter Rinck, PhD, is a professor of diagnostic imaging and the president of the Council of the Round Table Foundation (TRTF) and European Magnetic Resonance Forum (EMRF).Still, there is neither an easily explainable causality nor any unmistakable evidence of a straight connection between these numbers and a distinct pathology. There is no unique and unambiguous signature of malignancies or other pathologies in tissue relaxation times, be it in ex vivo or in vivo measurements. Many people believe that numbers -- or, as it is more fashionable to say, data -- are the truth but they do not understand how the numbers were acquired and what they stand for. Nature doesn't care about numbers. Believing in such postulations many years after they have been dismissed is a sign of scientific naiveté.
What's wrong in relaxation time mapping and applications is the precondition and presumption that a difficult biological structure, such as a tissue or tissue changes in the human body, can be quantified and qualified with crude nuclear magnetic resonance proton relaxation parameters.
Quantity and quality are being confused; it's so easy counting something -- which doesn't mean that one can classify or characterize with numbers what one counts. The components, chemical, and electrical processes in a tiny volume element, no matter how small it is, are far too complex and fickle to be expressed in bare figures. More so, on closer inspection, "objective" procedures, "objectively" defined range values, as well as "objective" quality indicators for measurements often prove to be biased and interest-driven.
In short, there is no precise numerical fingerprinting based on relaxation constants in biomedicine.
It is helpful to look into a microscope and see how complex and complicated tissue structures are, both in normal and in pathological tissues -- and in not-normal but not (yet) pathological tissues.
In the end, it is not necessarily the errors or procedural "confounders" connected to the most elaborate and sophisticated data acquisition that make typing of normal and pathological tissues or grading of diseases impossible, but rather the complexity of tissue composition and the overlapping of relaxation time values of heterogeneous volume elements examined and processed into a single number or number range.
Nowadays, lessons are rediscovered that became clear 25 years ago and finally admitted, though diplomatically beating around the bush:
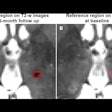
In conclusion, our question, whether native T1 mapping in cardiac MR imaging can differentiate between healthy and diffuse diseased myocardium, must be answered with "yes" and "no," because the native myocardial T1 relaxation time allows discriminating between groups of patients with certain diffuse myocardial pathologies and a group of healthy individuals, but does not allow differencing between healthy and diffuse diseased myocardium in individual subjects.5
Researchers also came to realize that novel methods for faster data acquisition deliver rough estimations but not accurate data. The higher the magnetic field, the larger seems to be the spread of T1 and T2 relaxation time estimations.
A vast extent of methods and sequences has been developed to calculate the T1 and T2 relaxation times of different tissues in diverse centers. Surprisingly, a wide range of values has been reported for similar tissues (e.g., T1 of white matter from 699 to 1,735 msec and T2 of fat from 41 to 371 msec), and the true values that represent each specific tissue are still unclear, which have deterred their common use in clinical diagnostic imaging.6
However, such indications are limited because increasingly different and simpler MR techniques exist that may lead to the wanted result.
In one of the next columns, I will try to discuss the nonscientific and nonmedical reasons why these measurements returned and why they will stay with us for some time.
Dr. Peter Rinck, PhD, is a professor of diagnostic imaging and the president of the Council of the Round Table Foundation (TRTF) and European Magnetic Resonance Forum (EMRF). He has no financial interests to disclose.
References
- Rinck PA. Relaxation times blues. Rinckside. 1991;2(1):5-7.
- Young I. Significant events in the development of MRI. J Magn Reson Imag. 2004;20(2):183-186.
- Rinck PA. Relaxing times for cardiologists. Rinckside. 2015;26(2):3-5.
- Rinck PA. MR fingerprinting returns to radiology -- and hopefully disappears again. Rinckside. 2015;26(5):13-14.
- Goebel J, Seifert I, Nensa F, et al. Can native T1 mapping differentiate between healthy and diffuse diseased myocardium in clinical routine cardiac MR imaging? PLoS ONE, 24 May 2016.
- Bojorquez JZ, Bricq S, Acquitter C, Brunotte F, Walker PM, Lalande A. What are normal relaxation times of tissues at 3 T? Magn Reson Imaging. 2017;35:69-80.
The comments and observations expressed herein do not necessarily reflect the opinions of AuntMinnieEurope.com, nor should they be construed as an endorsement or admonishment of any particular vendor, analyst, industry consultant, or consulting group.