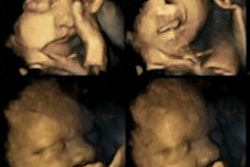
A new method of segmenting the fetal heart with echocardiography could improve the evaluation of congenital heart defects compared to the time-consuming manual segmentation in use today, according to researchers from the U.K.
Echocardiography is the method of choice for diagnosing fetal congenital heart defects, which occur in about one in 8,000 births. Measurement of absolute heart chamber size is required to evaluate the fetal heart. But unlike in adult patients, the walls separating the chambers in developing fetuses are too thin to be segmented automatically, particularly in the atrial septum, the membraneous segment of the ventricular septum, and the valve leaflets.
"The condition we're looking for is called immuno-hemolyis, this is where the fetal heart grows larger in order to compensate for an attack on the red blood cells" characteristic of the disease, said Irving Dindoyal, Ph.D., from University College London in the U.K. "Also fetal anemia ... can cause the heart to grow larger, so if you can detect this condition in the segmentation you can diagnose it in pregnancy."
Thin membranes separating the chambers can present a thorny problem for segmentation because ultrasound cannot usually distinguish them, said Dindoyal who spoke at the 2009 Computer Assisted Radiology and Surgery (CARS) meeting in Berlin.
What previous researchers have done is segment them manually, a process that is time-consuming and operator-dependent, he said, noting that in 2008 Tutschek et al used an ultrasound scanner with a proprietary algorithm to partially automate the segmentation process.
To segment the chambers automatically, the researchers developed a region-based deformable model based on level sets. They tailored the approach previously used in adult hearts to allow simultaneous segmentation in multiple segments when the membrane separating the chambers was extremely thin or absent.
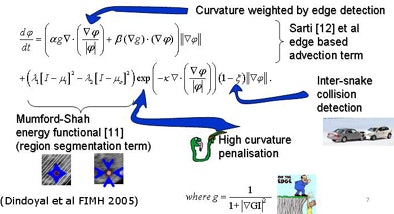 |
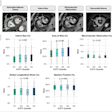
| Segmentation algorithm uses a deformable model based on level sets. Segmentation of multiple regions is performed simultaneously by placing a seed in the center of each chamber before initializing the process, which takes about 10 minutes. The deformable model grows from the initial seed point to fill the entire chamber. Any holes between chambers such as partially resolved atrial septal defects are reconstructed by the individual growing surfaces as they meet each other. The collision-awareness function halts the growth of intersecting surfaces and prevents interchamber leakage. All images courtesy of Irving Dindoyal, Ph.D. |
"You can imagine the deformable model as something that more or less wraps around the shape of interest you're trying to segment," acquiring a similar shape, he said.
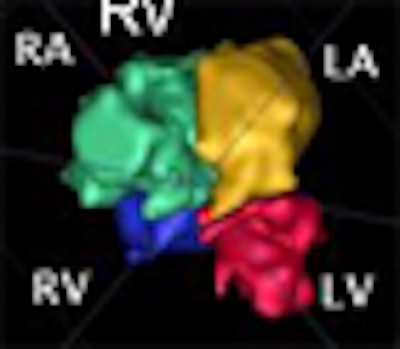
The chambers were distinguished using a "collision detection" method that the study authors developed in 2005, placing a seed point and then using region growing to fill out chambers simultaneously until the regions collided, eliminating the need for a visible wall separating the chambers, Dindoyal explained. The study compared the accuracy of 2D and 3D ultrasound representations of the fetal heart to manual segmentation.
"You place the seed points manually, one at the center of each chamber, and you place them only in one plane," he said. "We reslice the datasets to four-chamber views to make it easier for clinicians to place the sequence. Then you set the maximum iterations, press go ... and you find it's segmented in about 10 minutes in 3D, and you can segment all four chambers simultaneously."
The 2D datasets were acquired using an Acuson 128 XP 10 scanner (Siemens Healthcare, Malvern, PA) at 25 frames per second using a 4- or 5-MHz probe and approximately 0.25-mm pixel size resampled by the scanner. The 3D images were acquired on a Sonos 7500 scanner with Live 3D (Philips Healthcare, Andover, MA) with an X4 matrix probe operating at 2-5 MHz at 20 vps.
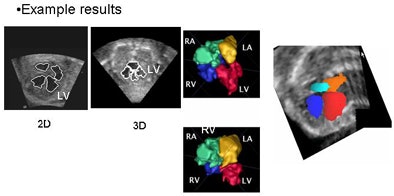 |
| In simultaneous chamber segmentation, seen here in 2D and 3D, collision detection awareness in the algorithm halts the growth of the intersecting surfaces and prevents interchamber leakage. The function works equally well in 2D and 3D datasets. |
"The computed error is basically the distance error between manual and automatic contours," applied to a 2D and a 3D dataset, with root mean square (RMS) error measurements performed by two observers, he explained. "From [the RMS error] you can get the average error, and then we computed the standard deviation across the sample range." Finally, the results were validated on a fetal heart phantom.
"The RMS error was under 1 mm for all datasets and across all cardiac chambers as well," Dindoyal said.
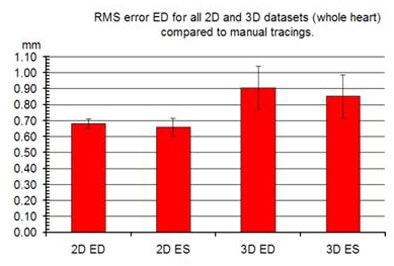 |
| Errors were calculated by computing the weighted root mean square (RMS) distance error from all chambers for a single dataset, then computing the standard error over all datasets. The error bars represent the standard error, which appears to show a larger error in 3D than 2D by a difference of less than 0.5 mm. In addition, the end systole (ES) segmentation is slightly better than the end diastole (ED), probably because in ED the valves are open and it is unclear where the atrial-ventricle boundary lies. The errors are less than 2 mm for all region-growing examples, meaning less than 10% considering the 20-30 mm length of the fetal heart. |
The algorithm will not replace manual segmentation, but it provides shorter time scales for segmentation, Dindoyal concluded. "It's very repeatable, and several sequences can be run at the same time," he said. The method is more accurate when seed points are placed roughly in the center of the heart chambers.
Manual tracings typically require a couple of hours just for a single cardiac phase and a 3D dataset, versus 10 to 15 minutes using the proposed method. In addition, several datasets can be initiated at the same time, he said.
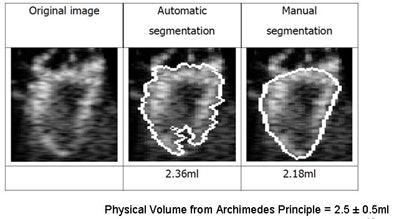 |
| Image results show volume calculations in manual versus automated segmentation (physical volume from Archimedes principle = 2.5 ± 0.5 mL). |
Automatic delineation is comparable to manual tracings of the fetal heart with RMS distance errors in 2D marginally lower than in 3D, Dindoyal concluded.
By Eric Barnes
AuntMinnie.com staff writer
November 6, 2009
Related Reading
Obesity, diabetes interfere with ultrasound detection of fetal anomalies, April 23, 2009
Early cardiac activity predicts good IVF outcome, December 23, 2008
CARS report: Fuzzy theory yields sharper CTA images, June 27, 2008
3D development outpaces facilities' readiness, June 23, 2008
Preliminary study finds fMRI possible in fetal heart, October 7, 2005
Copyright © 2009 AuntMinnie.com