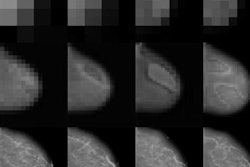
Unless radiologists have shut themselves off in a darkened room over the past few years, they will have heard about the incoming tsunami of artificial intelligence (AI) algorithms poised to make their jobs faster, their reports more precise, and their practice more impactful.
The motivated among us will have begun looking at how we can get involved with developing AI, and some, like me, will already be actively working on use-cases, research, and validation for specific tasks. This is great, but I guarantee whatever dataset you are working on isn't big enough and you are thirsty for more. Maybe you have access to the free 100,000 chest x-rays from the U.S. National Institutes of Health (NIH)? Or maybe you convinced your hospital to let you sniff around its PACS? Or maybe you work for IBM and have access to all that sweet Merge healthcare data?
 Dr. Hugh Harvey.
Dr. Hugh Harvey.Good for you. The hustle to get access to medical data and make it exclusive and proprietary reminds me of the gold rush of two centuries ago. Prospectors arriving in droves, bargaining with investors for capital to start digging, begging for finance to buy up plots of land, and pitching for investment for equipment while promising fortunes in gold. However, there is an unspoken and too often ignored hurdle to jump before digging in to the really juicy stuff and pulling out chunks of solid ore.
The problem is medical imaging data isn't ready for AI.
Developing machine learning algorithms on medical imaging data is not just a case of getting access to it. While access is of course a huge headache in itself -- look at DeepMind for a clear example -- it is not the only hurdle in the race. In this article I will explain the concept of data readiness, and how investment in preparing your data is more important than developing your first algorithm.
Stages of data readiness
There is no defined and universally accepted scale to describe how prepared data is for use in AI. Many management consultancy firms will offer pretty slide shows on how you can gather data and create insights from it -- and charge through the nose for the privilege -- but the underlying principle of data readiness still remains a largely unquantifiable process.
A little known but thoroughly important paper on data readiness by Neil Lawrence, professor in computational biology at the University of Sheffield in the U.K., caught my eye. After reading it and contacting him to discuss it, I came up with my own modified version of his data readiness scale (see below).
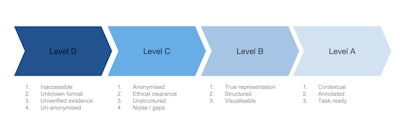
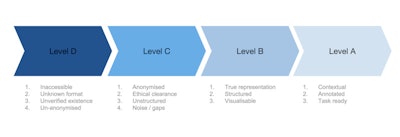
Medical imaging data readiness scale.
Let's start at the beginning, with level D.
Imagine you own a massive oil field -- a pretty nice thing to imagine. The problem is, your oil field is totally inaccessible to the people who want to dig up that oil and turn it into petrol and make you huge profits. Not only that, there are laws and ethical blockades in the way. Nobody, not even you, is sure what exactly is in your oil field. Your oil field is what I call "level D" data.
Level D data is that which is unverified in quantity and quality, is inaccessible, and is in a format that makes it difficult or impossible for machine learning to do anything. This level of unanonymized data is in every hospital's PACS archive in massive volumes, just sitting there, doing nothing, except acting as a record of clinical activity. (And, I shudder at the thought, every so often due to data storage issues, some U.K. National Health Service (NHS) trusts, or hospital groups, actually delete backlogs of this data. Like throwing away oil.)
In order for level D data to be distilled up into level C, you need to build a refinery. The first stage of data refining is to get ethical clearance for access to your data. In practice, this is done through a data-sharing agreement either locally with yourself via an ethics committee, or with a third party (university, company, or start-up).
NHS trusts, for example, may have thousands of data-sharing agreements in place at any one time. These agreements will also include clauses on data anonymization, as obviously no one wants the NHS to give away confidential patient information. So far so good. However, the data is still very unstructured, and will not be representative of a complete set. It will also be extremely noisy, full of errors, omissions, and just plain weird entries. Whoever has access to the data now has to figure out how to make it useful before they can get cracking on algorithmic development; C level data is ready to be given out to AI developers, but is still far from ready for being useful.
Let it B
The data now need to be refined further into level B data, by structuring it, making sure it's representative of the data you think you have, and running visualizations on it to be able to draw out insight into noise characteristics and other analysis metrics. This process is actually even harder than the D to C stage, as it is bespoke for each dataset. There is no standard way of checking medical imaging data, and each individual group that has access to your data will be running their own data visualization and analyses. This is because data from different hospitals will have different characteristics and be in different formats (e.g., different DICOM headers, date, and time stamps, etc.). The process of converting level C to B can take months -- not exactly what researchers or start-ups need in the race for gold. Only with level B data can you have an idea of what is possible with it, and where AI can be used to solve actual problems.
Level A data is that which is as close to perfect for AI algorithmic development; it is structured, fully annotated, has minimal noise, and, most importantly, is contextually appropriate and ready for a specific machine-learning task. An example would be a complete dataset of a million liver ultrasound exams with patient age, gender, fibrosis score, biopsy results, liver function tests, and diagnosis all structured under the same metatags, ready for a deep learning algorithm to figure out which patients are at risk of nonalcoholic fatty liver disease on B-mode ultrasound scans.
Annotation is perhaps the hardest part in radiology dataset refining. Each and every image-finding exercise should ideally be annotated by an experienced radiologist so that all possible pathologies are highlighted accurately and consistently across the entire dataset. The problem is that barely any of the existing medical imaging data anywhere in the world is annotated in this way. In fact, most images in the "wild" aren't even annotated. That's why there is an entire industry centered on data tagging.
Ever signed into a website and been asked to click on images that contain road signs or cars? You're tagging data for self-driving car algorithms! Of course, not every internet user is a radiologist, so this crowdsourcing model doesn't work for medical imaging. Instead, researchers have to beg or pay for radiologist time to annotate their datasets, which is a monumentally slow and expensive task (trust me, I spent six months during my thesis, drawing outlines around prostates). The alternative is to use natural language processing (NLP) on imaging reports to word-mine concepts and use them to tag images, but this model is far from proven to be robust enough (yet).
Speculate to accumulate
The above refining processes to get level D data all the way to level A is expensive, both in terms of time and resources. All too often I see small research groups start out with a bold proposal to solve a particular task using machine learning, hustle to get data access, get stumped as to why their algorithms don't ever reach a useful accuracy percentage, and then give up. They tried to avoid the refining process and got caught out. Even the big players, like IBM, struggle with this problem. They have access to huge amounts of data, but it takes inordinate amounts of time and effort to prepare that data to make it actionable and useful.
As with any speculative investment, you have to finance infrastructure development to reap benefits downstream. That's why I have proposed a national framework for undertaking this work, which I call the British Radiology Artificial Intelligence Network (BRAIN).
The idea is to use the recently announced U.K. government life sciences strategy to secure funding to create an entirely new industry based on the oil field of NHS imaging data. BRAIN would act as a "data trust" or warehouse for anonymized imaging data, allowing researchers and companies access in return for equity in intellectual property (IP). By setting up BRAIN, the NHS would immediately bring data readiness to level C, and then subsequently each and every research activity would help refine the pooled data, moving the data up from level C to level A, creating a rich resource for the development of AI, adding value to the dataset.
The advantages of such a setup are huge. Not only does the NHS open up immense amounts of data while maintaining control, it also gets to benefit from the returns of AI development twofold by having its data refined and having a share of any IP produced. In addition, NHS hospitals no longer need to manage thousands of data-sharing agreements, which is a resource-intensive task. They simply point people toward BRAIN.
The benefits to researchers and companies are also significant: They all get access through a centralized data-sharing agreement (no more hustling individual hospitals one by one); the pool of data is larger than anything they previously had access to; and, over time, they spend less and less resources on data refining and more time on developing useful algorithms.
The benefits don't stop there. By pooling data from across the NHS, you reduce biases in the data, and expose algorithms to a wider range of imaging techniques, reporting styles, and pathologies, ultimately making the end-product algorithms more generalizable and available for use in any setting.
Growing a new industry
Metcalfe's Law states the value of a network grows as the square of the number of its users increase; just look at Facebook or Twitter, both massive networks with huge value. By centralizing access to a large pooled dataset and inviting hundreds of researchers to play, you immediately and dramatically increase the value of the network. All it takes is a commitment setting it up. We only need to look at the success of the UK Biobank to get an inkling of why such a network is crucial (and they only have just over 1,000 MRI exams).
 Metcalfe's Law and Moore's Law have relevance to AI today.
Metcalfe's Law and Moore's Law have relevance to AI today.The imaging AI landscape is like the Wild West -- and it needs a sheriff in town. By pooling structured data, we can create a new industry in research, while reaping the rewards. This is my big dream for massive data in medical imaging and AI; I hope I've convinced you to share in it!
Dr. Hugh Harvey is a consultant radiologist at Guy's and St. Thomas' Hospital in London. He trained at the Institute of Cancer Research in London, and was head of regulatory affairs at Babylon Health, gaining the CE Mark for an AI-supported triage service. He is a Royal College of Radiologists informatics committee member and adviser to AI start-up companies.