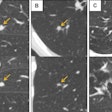
Like a human locked in a white soundless room struggling to make sense of the outside world, artificial intelligence (AI) is impossible to achieve without some sort of input. Even the best machine-learning engineers in the business can't make a functional algorithm without any data.
The human brain is constantly bombarded with data -- visual, auditory, tactile, olfactory, emotional, and more -- and has evolved advanced filters to sort through this onslaught to allow only meaningful inputs through. If we are to achieve a true AI, then the key lies in data, and the filtering and processing of it in vast quantities. However, right now, when it comes to training models for applications in medicine, there just doesn't seem to be enough high-quality data available to researchers.
Current frameworks for training artificially intelligent systems for radiology largely rely on supervised and semisupervised learning, where large datasets containing expertly labelled images are used. Researchers face three significant hurdles in obtaining the good stuff -- quantity, quality, and ethical use of medical data.
In addition to these hurdles, the time and cost it takes to climb the Medical Imaging Data Readiness (MIDaR) scale to convert level D data (live unanonymized clinical data inside hospitals) to level A (de-identified, labeled, clean data for training) can be a matter of several months, man-hours, and many thousands of dollars. Even when level A data are obtained, it is often biased and imbalanced, despite best attempts to create the perfect dataset. Rare diseases will be underrepresented, and all sorts of weird and wonderful normal variations will abound.
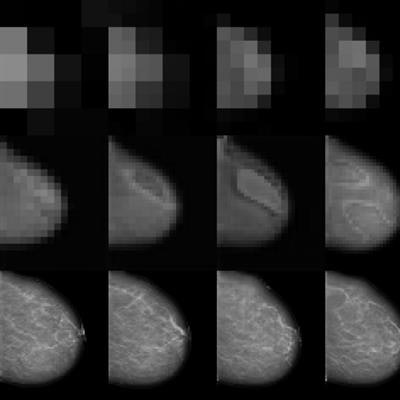
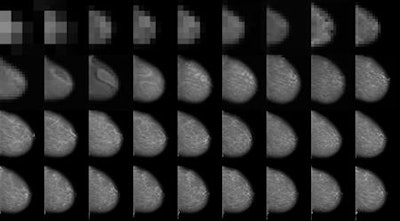 Image taken from high-resolution mammogram synthesis using progressive generative adversarial networks. Courtesy of Korkinof et al., Kheiron Medical Technologies, https://arxiv.org/abs/1807.03401.
Image taken from high-resolution mammogram synthesis using progressive generative adversarial networks. Courtesy of Korkinof et al., Kheiron Medical Technologies, https://arxiv.org/abs/1807.03401.Very few publicly available high-quality medical imaging datasets are available for research, and no globally standardized datasets for validating radiology AI model accuracy are yet available. This problem is most evident in the imaging sector, as opposed to electronic health record analysis for instance, where data are largely tabulated and alpha-numeric. This type of data can fairly easily be augmented, simply by generating random numbers for dates, or using more sophisticated techniques for generating fake patient profiles.
In medical imaging, techniques for data augmentation such as flipping, cropping, zooming, and skewing images can only go so far. After all, there's only so many ways you can alter an image, and even then, no new data are actually added.
Unfortunately, you can't generate infinite synthetic medical images to train on...or can you?
Creating realistic fakes
Machine learning teams across the globe are now turning their attention to creating synthetic medical imaging data, using a relatively new form of network (first introduced by Goodfellow et al. in 2014) known as a Generative Adversarial Network (GAN). This trend is accelerating, with an increasing number of papers on the generation of medical images coming out.
You are likely to have seen GANs in action already in other sectors. They are famous for creating images of fake celebrities, for instance, as well as fake videos of people dancing like pros.
 Examples of generated realistic high-resolution images of fake celebrities.
Examples of generated realistic high-resolution images of fake celebrities.So what are GANs, and how do they work?
GANs are comprised of two networks, one generative (G) and one discriminative (D). I like to think of them as a pupil and a teacher respectively. They function against each other (hence the term adversarial), with the pupil trying to pass a test and the teacher marking the test. The test happens to be whether or not the pupil can create a realistic looking synthetic image. Only when the pupil gives a correct answer does the teacher reward the pupil. Importantly, both the teacher and the pupil have little or no prior knowledge of the syllabus. The teacher must learn iteratively from real images and "grade" the pupil, and the pupil must guess over many iterations what the answers might be, which is the far harder task!
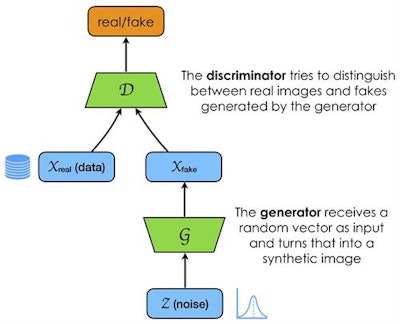 Flow of data.
Flow of data.In this way, the pupil learns in an unsupervised manner -- it has not been explicitly taught anything, and only gets feedback on whether it is right or wrong.
Generators for medical images must use a random noise vector input to create synthetic images that look real enough so a discriminator recognizes them as belonging to the distributed feature (or latent) space that defines the task at hand. It can be tricky to conceptualize the latent space found within a dataset of images, as it is essentially a multidimensional abstract entity. In fact, in order to narrow down this feature space researchers usually constrain it to lie on the surface of multidimensional hypersphere.
Multidimensional hyperspheres
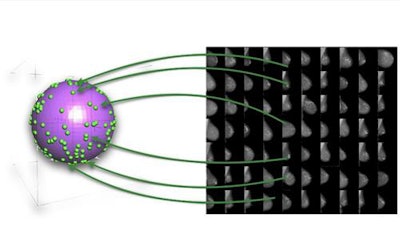 A multidimensional hypersphere with discrete points relating to the feature space within images.
A multidimensional hypersphere with discrete points relating to the feature space within images.Imagine a sphere in a 3D space (x, y, z) and discrete points lying on its surface. Each point represents a synthetic image and as the generator is trained from the discriminator's feedback, the points on the sphere gradually encode more and more information about the features found in real-word medical images. For instance, certain directions on the sphere may have semantic meaning, which in the case of mammography may relate to density or parenchymal pattern. Assuming a perfectly trained generator, each point would represent the entire set of features found in real-world medical images necessary to construct them synthetically. In reality this space isn't actually 3D, but rather multidimensional (a hypersphere), nevertheless, its dimensionality is much smaller (more compact) than the images, which can consist of millions of pixels.
The discriminator network (D) encodes the images into its own high-dimensional representation, similar but not necessarily aligned with the generator's (G). It absorbs knowledge about contents and boundaries of the feature space of a given training dataset, while the generator network (G) attempts to generate new synthetic points to fit within the same space. If the generated images don't fit within the feature space boundaries, the discriminator rejects them. Through multiple iterations the generator network learns to create images that start to fit, and does so with increasing accuracy and completeness. The result is a feature space full of infinite discrete synthetic data points, all fitting within the feature/latent space of the original dataset. What's more, potentially none of the synthetic images are identical to any of the original training set. They just share the same distribution in the feature space.
 Dr. Hugh Harvey.
Dr. Hugh Harvey.If that was difficult to wrap your head around (it took me a few tries), imagine a simpler task -- teaching a child to draw a carrot. First, you show them a carrot and ask them to draw it. Chances are they will draw something vaguely resembling the carrot you've shown them. What they've done subconsciously is to "learn" the feature space of what it takes to be a carrot -- orange pointy thing with green leaves at the top. Now imagine showing the child ten different carrots, and then ask them to draw a new picture of a carrot. They will be able to abstract and draw an entirely new carrot, one that is different to all the pictures you've shown them, but still entirely recognizable as a carrot. Humans are exceptionally good at this sort of imaginative and creative task.
And this is why GANs are so interesting -- they take us a few steps closer to replicating how the human brain actually works. We humans are capable of abstract and playful thought, we don't learn just by parroting. Humans learn in an unsupervised way, and as children we experiment and play to learn about our environments, receiving constant feedback. In a way, we learn by creating synthetic realities in our subconscious and play-testing scenarios out in our heads. GANs in essence are capable of doing the same thing. By creating abstract concepts, and ensuring that they fit within certain predefined parameters, machines can effectively be made to hallucinate, within the hallucinations remaining within the limits of reality. It is visual expression, creation and abstraction, and may be a huge step forward in unlocking true artificial intelligence machine vision.
So why are GANs useful?
There are several proposed use cases for synthetic medical imaging data. I won't cover them all, but I urge readers to refer to this excellent review paper by Kazeminia and colleagues for a more complete and technical insight.
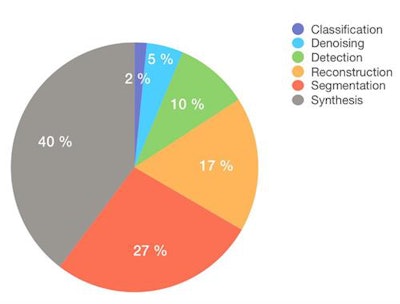 The proportion of GAN research in different areas.
The proportion of GAN research in different areas.Image synthesis remains the most studied application. For instance, the team at Massachusetts General in Boston, U.S., published their work on generating synthetic MRI brain scans, and others have produced realistic retinal images. There has also been extremely interesting research on the de-noising of images (improving low dose CT resolution), image reconstruction from sparsely acquired data (faster MR sequences), as well as attempts at improving segmentation and elastic registration algorithms.
Due to computational and processing power limitations, there is a technical barrier to creating very high-resolution images, with most current research producing images between 124 x 124 and 512 x 512. This is in part because GANs are notoriously fragile, often collapsing and producing rubbish as the layers within the networks become overcrowded.
At Kheiron, we recently published our work on generating high-resolution mammograms using what is known as a progressive GAN (or pGAN). By training a discriminator network on around half a million real mammograms from our database, we mapped the entire feature space of what constitutes a mammogram, and then trained a generator to synthesize high-resolution synthetic images within the same feature space. This was done progressively (see picture at top of page), starting both discriminator and generator at very low resolutions, and gradually doubling the image size at each iteration.
To our knowledge, we exceeded the highest resolution for synthetic 2D medical images published to date, managing to create extremely realistic mammograms at a pixel ratio of 1280 x 1024. (As resolution doubles, the feature space becomes almost exponentially more complex!). In addition, we have since been able to generate both standard views (craniocaudal and medial lateral oblique) of each breast simultaneously, meaning that we can create entire synthetic patient cases of any shape, size, or density of breast, not just single images which are unrelated. Furthermore, we are able to generate an infinite amount...as long as the generated images remain within the trained feature space. The video below demonstrates how realistic these synthetic images look, and how the entire feature space can be passed through.
This is early research, and the full ramifications of this work are not yet known. Not all the generated images look absolutely real -- there are certainly some odd cases that limit the usefulness.
We will be looking into whether or not the realistic synthetic images can be used to augment datasets for downstream classifier training. It is worth noting that synthetic images, while infinite in number, do not represent "new" data outside of the boundaries of the original dataset latent space, and therefore cannot be used as a substitute for training on high-quality real-world data. However, by using synthetic data to train as weakly labeled data prior to fine-tuning on well-annotated real data, there may be considerable benefits to be reaped.
The discriminator may be useful as a detector for real images that are out of the normal feature space distribution. There is also work to be done to assess whether these models can aid in domain transfer, particularly for normalizing the appearance of mammograms taken from different vendor hardware.
So, while it's not quite "free infinite data" just yet, we have certainly taken one step toward getting there by letting artificial intelligence get infinitely creative. After all, that's how we humans learn.
Dr. Hugh Harvey is a Royal College of Radiologists (RCR) informatics committee member, clinical director at Kheiron Medical, and advisor to AI start-up companies, including Algomedica and Smart Reporting. He trained at the Institute of Cancer Research in London, and was head of regulatory affairs at Babylon Health.
The comments and observations expressed herein do not necessarily reflect the opinions of AuntMinnieEurope.com, nor should they be construed as an endorsement or admonishment of any particular vendor, analyst, industry consultant, or consulting group.