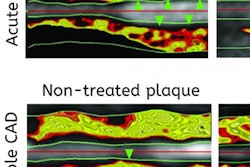
VIENNA - Automated plaque analysis in coronary CT angiography can predict which patients with known ST-segment elevation myocardial infarction (STEMI) may go on to have a heart attack in the future, concluded a Wednesday presentation at ECR 2016.
The small study used investigational plaque analysis software to quantify different plaque types based on CT attenuation. Investigators then sleuthed out patterns associated with future cardiac events in their CSI -- or "coronary scene investigation" -- to determine the cause of the later MI, said presenter Dr. Stefanie Mangold from the Medical University of South Carolina (MUSC) in the U.S. and the University of Tübingen in Germany.
 Dr. Stefanie Mangold from MUSC and the University of Tübingen.
Dr. Stefanie Mangold from MUSC and the University of Tübingen."The software evaluated the plaque composition, and analysis of plaque composition may help predict cardiac events," Mangold explained in her talk, cautioning that the results need to be verified in a larger population.
The study, led by Stephen Fuller from MUSC, examined the use of pre-STEMI cardiac CT in order to define predictors of future cardiac events.
In all, 28 patients underwent coronary CT angiography on a dual-source CT scanner (Somatom Flash, Siemens Healthcare); however, the exclusion of patients who had stents or poor image quality left just 13 who underwent automated plaque analysis using an investigational software package (syngo.via Frontier, Siemens). Still, the results of plaque analysis showed a rather homogeneous pattern of culprit lesions in the mid right coronary artery (RCA).
Two experienced readers identified the culprit lesions in consensus, and plaque analysis was performed on a per-segment, per-vessel, and per-culprit-lesion basis. Culprit plaque composition was evaluated using mean attenuation thresholds, based on a 2012 meta-analysis:
- Fibrotic: 71-124 HU
- Lumen: 125-511 HU
- Calcified: > 511 HU
"The software gives you the total volume of the segment, the volume of the lumen, and the volume of the plaques, and you can calculate the lumen narrowing in percentage," Mangold said.
Results of the per-vessel and per-segment analyses showed mean vessel lumen reductions of less than 50%. In all segments, the lipid-rich component comprised only 21% of plaque volume, while the most prevalent plaque component was fibrotic, at 71%.
The mean lumen reduction in the coronary arteries was only 23% in the overall evaluation, but lumen narrowing nearly doubled to 45% (p < 0.01) in the culprit lesions. The lipid component was also substantially higher, at 45%, compared with 21% in the overall analysis.
"Evaluating the culprit plaque showed the main localization was in the right coronary artery with a prevalence of 71%, and the average time for the adverse event was around three years after coronary angiography," Mangold said.
| CT results at mean 2.9 years before MI | ||
| Overall plaque analysis | Culprit-plaque analysis | |
| Volume reduction | 23% | 45% |
| Calcification | 5.9% | 0.8% |
| Lipid-rich | 21% | 45% |
| Fibrotic | 71% | 54% |
"The lipid-rich component of culprit plaques is increased compared to plaques from all segments in a STEMI population," Mangold concluded. The software identifies plaque composition, which may predict future cardiac events, though the results are preliminary and must be validated in a larger population, she added.
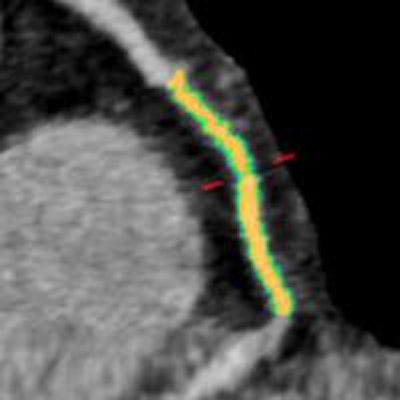
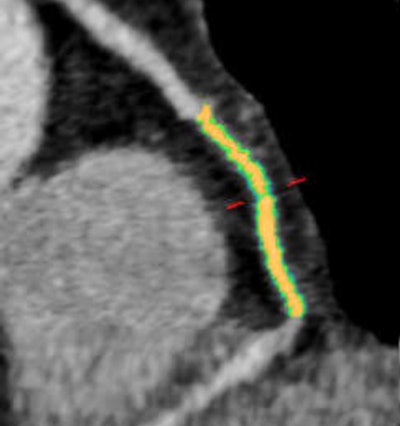 Woman age 61 years with a history of arterial hypertension, hyperlipidemia, and an abnormal stress test. Initially, CT angiography revealed patent coronary arteries and no significant stenosis, but after the patient had a myocardial infarction, a retrospective plaque analysis showed a 75% reduction in lumen volume in the mid right coronary artery; the culprit plaque had a lipid component of approximately 60% and a fibrotic component of about 40%. The mid right coronary artery was the most common location for culprit plaques in the patient cohort. Image courtesy of Dr. Stefanie Mangold and Stephen Fuller.
Woman age 61 years with a history of arterial hypertension, hyperlipidemia, and an abnormal stress test. Initially, CT angiography revealed patent coronary arteries and no significant stenosis, but after the patient had a myocardial infarction, a retrospective plaque analysis showed a 75% reduction in lumen volume in the mid right coronary artery; the culprit plaque had a lipid component of approximately 60% and a fibrotic component of about 40%. The mid right coronary artery was the most common location for culprit plaques in the patient cohort. Image courtesy of Dr. Stefanie Mangold and Stephen Fuller.Session moderator Dr. Filippo Cademartiri from the Montreal Heart Institute asked if it was difficult to obtain accurate plaque assessments given that the regions of interest selected for analysis might contain vessel wall or small areas of several kinds of plaque that could confound the analysis.
"On the CT alone, it's very difficult to be sure of what we name, if we name something lipid or fibrotic or average or mixed, or even calcified when they are small calcifications," he said. "Did you find it difficult to segment noncalcified plaque, especially in the right coronary artery where you have fat surrounding the artery?"
A lot of cases were excluded due to motion artifacts, "and we only used the studies where the image quality was sufficient and the composition of the plaques could be identified," Mangold responded.